next steps
EXPAND PIPELINE
- MRI Analyzer’s pipeline is currently automated and only requires specifying an input folder that contains the training dataset
- Incorporating other cancer types with new copies of MRI Analyzer’s pipeline can broaden its usefulness
- Additional datasets:
- Prostate Cancer: PRAD-CANADA – 392 subjects
- Breast Cancer: Breast Cancer Screening-DBT – 985 subjects
- Colon Cancer: CT Colongraphy (ACRIN 6664) – 825 subjects
- Lung Cancer: NSCLC-Radiomics – 422 subjects
- Kidney Cancer: C4KC- KiTS – 210 subjects

ADD OBJECT SEGMENTATION via yolo5
Because each medical scan contains several slices, each of which may or may not include cancer, current processing for computer vision models requires:
- Creating a single label for each scan
- Manually cropping each image slice to only the interior portion, which is centered on the prostate
- Dropping the first and last sets of image slices, which contain no / the least amount of prostate
- Combining the remaining image slices into a collage
Object segmentation can be incorporated into the pipeline via the YOLO5 model to improve processing. The updated pipeline would only require:
- Inputting a single MRI scan
- Extracting the prostate from within each slice, if present, automatically via the trained YOLO5 model
- Combining the extracted prostate images into a higher resolution, less noisy collage
This would have the advantage of providing more relevant information into each computer vision model, likely increasing performance.

CANCER SLICE CLASSIFICATION
The original dataset provided cancer information in the form of biopsy results for each patient, which was used to generate the cancer / no cancer label.
In addition, 3-D coordinates were also provided which mapped each biopsy sample to its location in the patient’s corresponding MRI image.
Incorporating this information in a useful way for either image pre-processing, or as a feature into the computer vision models, will allow more detailed learning of how image features relate to cancer outcomes. This is a unique feature of the current dataset and will likely increase performance.
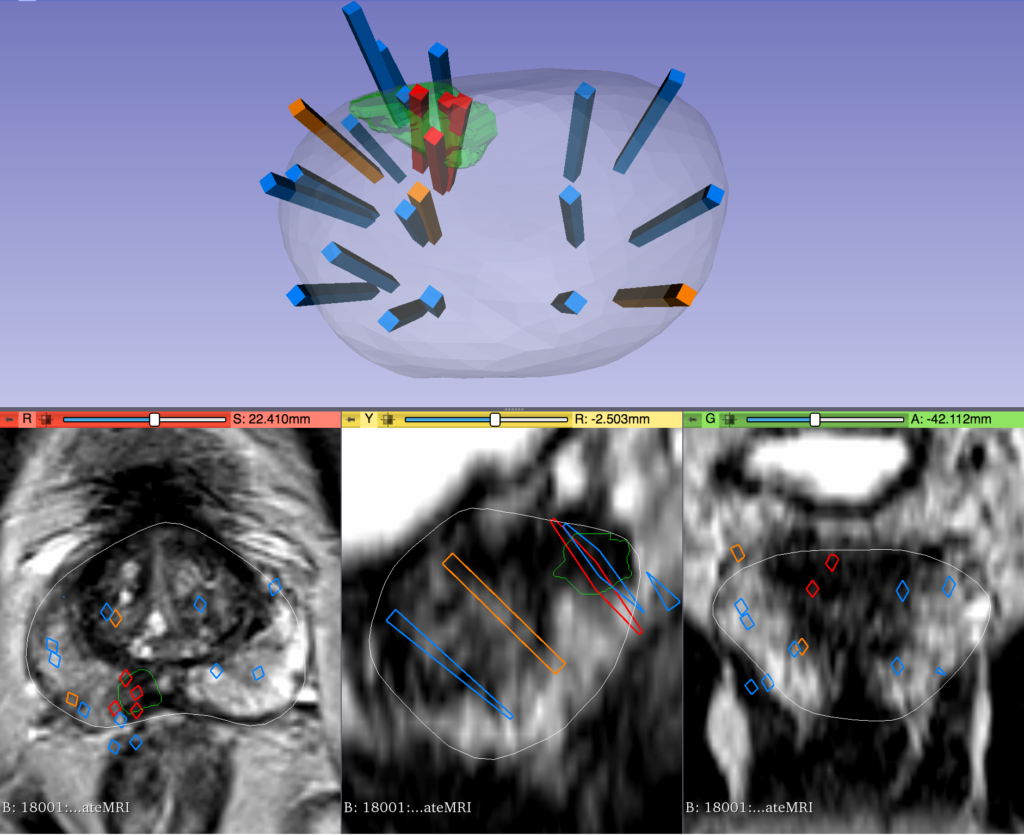
IMAGE CHANNELS & OVERCOMING HARDWARE LIMITATIONS
Images that have been used for this study were put into a single channel collage due to hardware memory limits. For the same reason, it was also required to lower image resolution within the collage. Both of these adjustments likely affected model training and performance. With more powerful hardware from future funding, images could instead be processed in a multichannel format corresponding to the physical location within the body. This could also provide the opportunity to keep images at higher resolutions where more details are available to the model. Each of these changes would likely increase performance as more information is available to the computer vision models.

related research
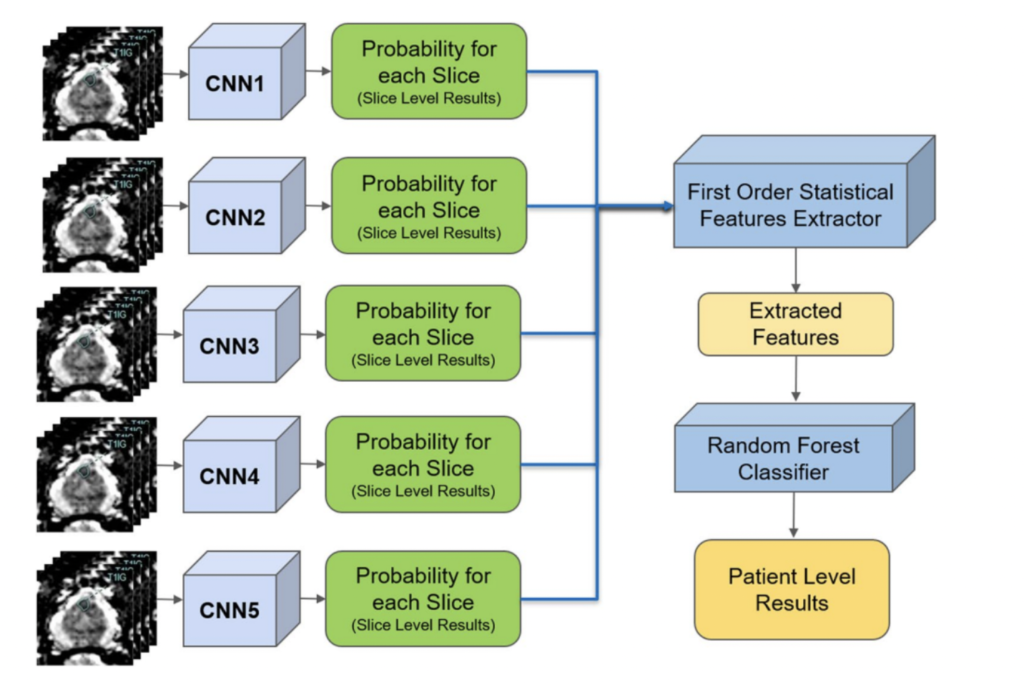
In a separate application of ResNet to prostate cancer detection (4), diffusion-weighted MRI images were used as an input with 319 training and 108 testing patients. Five stacked ResNet17 models were used in an ensemble in order to make slice-level classifications, which were then used to generate a single patient level classification. This is shown in the image on the right.
The reported AUC-ROC was 0.84 – 0.90 at the slice level and 0.76 – 0.91 at the patient level. Slice level indicates a prediction of whether or not an individual MRI slice had a prostate cancer tumor, while patient level indicates whether an entire MRI scan for a patient had a prostate cancer tumor.
In a statistical study that was conducted by Smith RP, et.al. (2), researchers analyzed historical data to see whether prostate cancer could be detected and diagnosed accurately based on the PSA (Prostate Specific Antigen) results. Specifically, they compared the PSA results of 977 patients with confirmed prostate cancer diagnosis against different clinically significant PC thresholds. The research concludes that the almost two thirds of the patients were diagnosed as having a prostate cancer based on the aligned results of the biopsy data and the PSA screening results.
Furthermore, in a study by Sonn et. al. (3), individual radiologists were assessed for how they evaluated MRI scans for prostate cancer. Nine radiologists provided a PIRADS score for different MRIs, which is a procedural metric when evaluating an MRI. The scores were used in a logistic regression in order to predict the clinical significance of cancer. Results showed both the PIRADS score distribution as well as the yield of clinically significant cancer varied across individuals. The research also found that the variability amongst radiologists was not due to factors such as education, training levels, etc.
Finally, in an article by Minh Hung Le et al. (1), researchers created a CNN and SVM ensemble to distinguish between clinically significant and insignificant prostate cancer from 364 patients. With a small dataset, they explored different data augmentation methods in order to improve results. It was found that fusion of CNN features did not guarantee an improvement in results. As a result, they used a new fusion method to create a feature learning process where data is more consistent with less variety. The results showed that the new fusion model demonstrated better performance compared to the other two methods also attempted in the study.
References:
- Le, M. H., Chen, J., Wang, L., Wang, Z., Liu, W., Cheng, K.-T. (T., & Yang, X. (2017). Automated diagnosis of prostate cancer in multi-parametric MRI based on multimodal Convolutional Neural Networks. Physics in Medicine & Biology, 62(16), 6497–6514. https://doi.org/10.1088/1361-6560/aa7731
- Smith, R. P., Malkowicz, S. B., Whittington, R., VanArsdalen, K., Tochner, Z., & Wein, A. J. (2004). Identification of clinically significant prostate cancer by prostate-specific antigen screening. Archives of Internal Medicine, 164(11), 1227. https://doi.org/10.1001/archinte.164.11.1227
Sonn, G. A., Fan, R. E., Ghanouni, P., Wang, N. N., Brooks, J. D., Loening, A. M., Daniel, B. L., To’o, K. J., Thong, A. E., & Leppert, J. T. (2019). Prostate magnetic resonance imaging interpretation varies substantially across radiologists. European Urology Focus, 5(4), 592–599. https://doi.org/10.1016/j.euf.2017.11.010
Yoo, S., Gujrathi, I., Haider, M. A., & Khalvati, F. (2019). Prostate cancer detection using deep convolutional neural networks. Scientific Reports, 9(1). https://doi.org/10.1038/s41598-019-55972-4