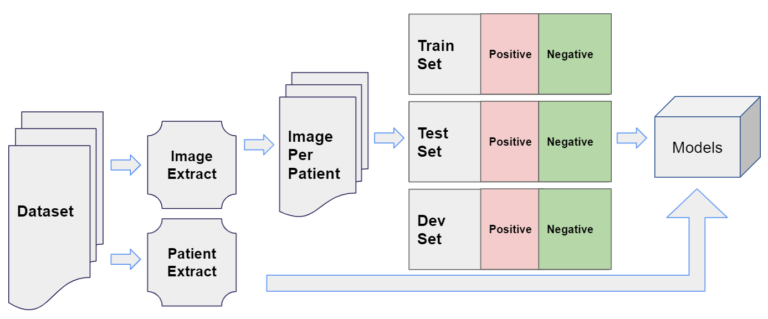
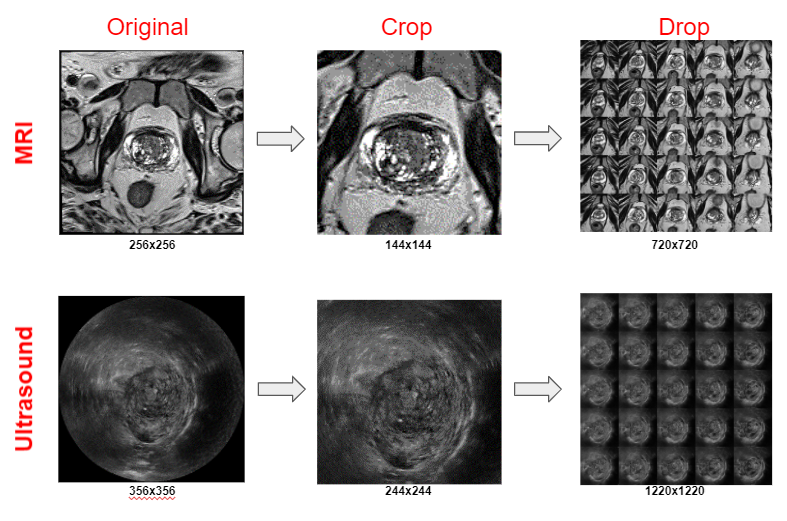
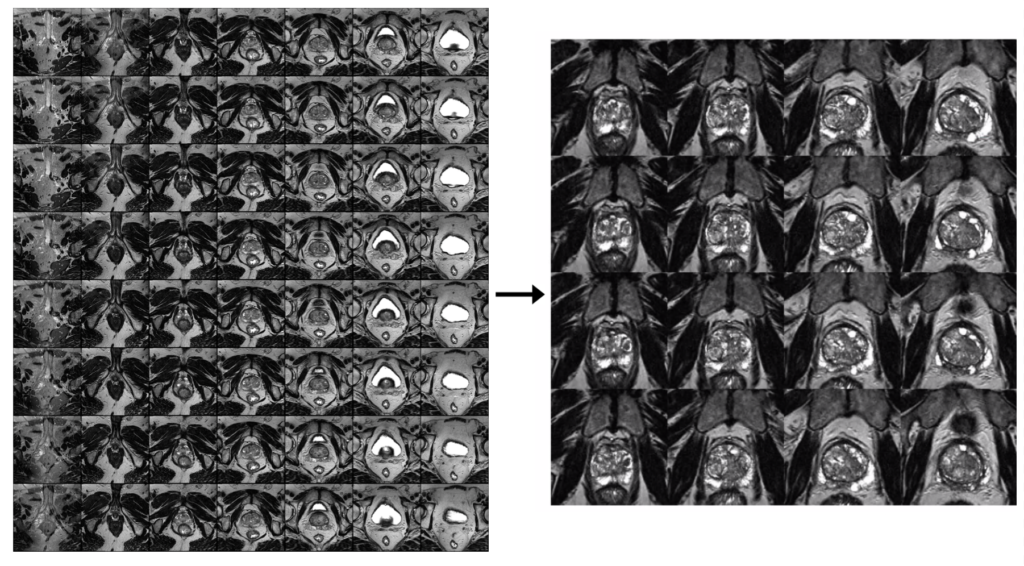
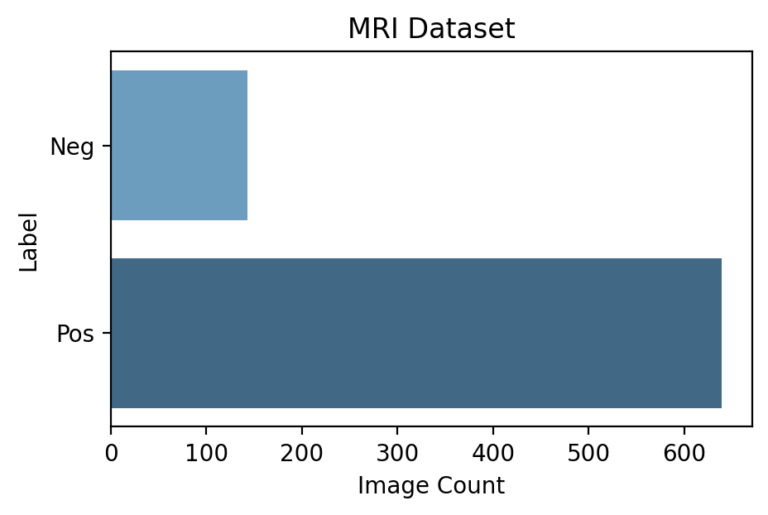
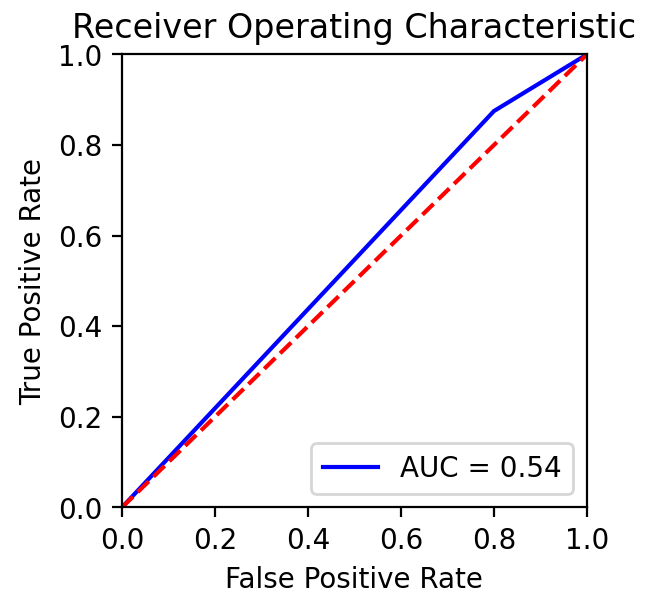
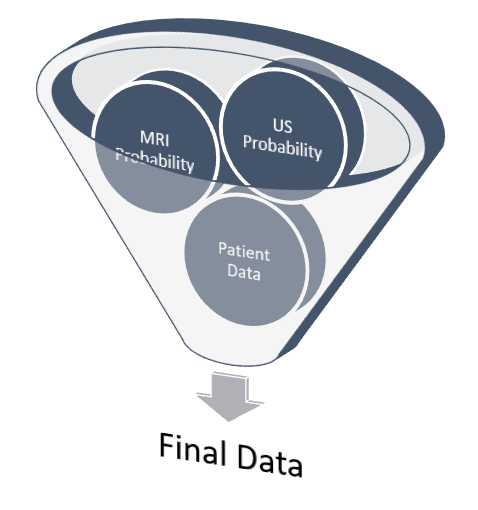
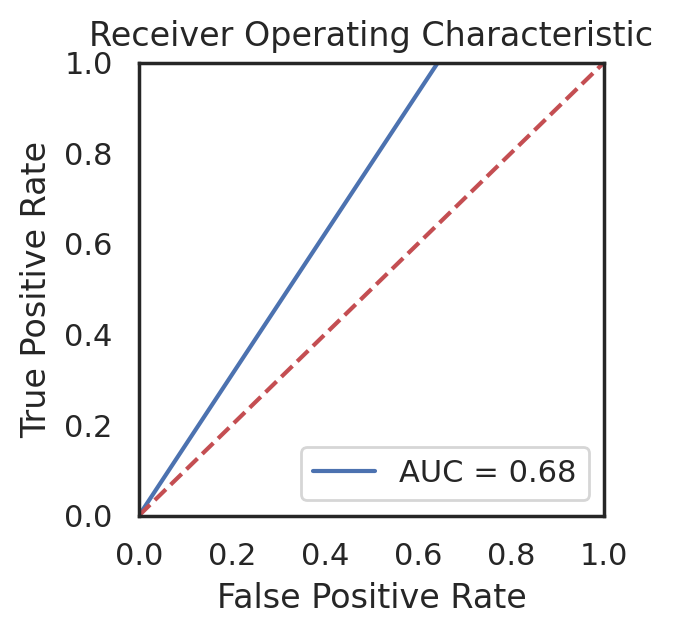
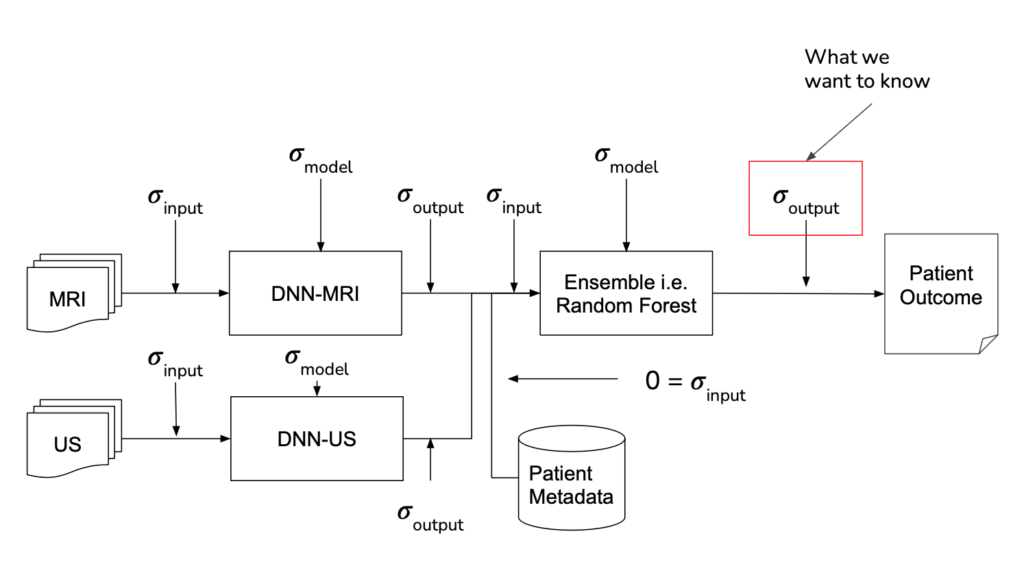
Project Environment
The environment used to create and run the algorithms for the MRI Analyzer are on the public cloud- Amazon Web Services. We are running an EC2 instance and within that running the Deep Learning for Ubuntu operating system. Within that, the docker image is based on NVIDIA for PyTorch. The results and dataset are stored in an S3 bucket and we utilized Papermill to run our models.